Picture-Perfect Memories: 5 Tips for a Successful Professional Family Photoshoot

Image Source- Google
Capturing family moments through professional photography is a wonderful way to preserve memories for a lifetime. Whether it's a family reunion, a holiday celebration, or simply a fun photoshoot, experienced family photoshoots can be cherished for years to come. To ensure that your family photoshoot is a success, consider the following tips:
1. Choose the Right Photographer
Look for Experience and Style
- Research photographers in your area and review their portfolios to get an idea of their style.
- Choose a photographer who specializes in family photography and has experience working with children and large groups.
- Consider the photographer's personality – they should be able to make your family feel comfortable and relaxed during the shoot.
Discuss Your Vision
- Communicate your ideas, preferences, and any specific shots you want with the photographer beforehand.
- Share any themes, locations, or props you have in mind to ensure your vision is captured in the photos.
- Work together to create a shot list to make sure all important moments are captured during the session.
2. Coordinate Outfits and Colors
Choose a Color Palette
- Select a color palette that complements each other and the overall theme of the photoshoot.
- Avoid matching outfits and instead opt for coordinating colors to add visual interest to the photos.
- Consider the location and season when choosing colors to create a cohesive look.
Dress for the Occasion
- Choose outfits that reflect your family's style and personality while ensuring everyone is comfortable.
- Avoid busy patterns or logos that can be distracting in photos.
- Accessorize with hats, scarves, or jewelry to add texture and dimension to the photos.
3. Plan Ahead
Schedule Wisely
- Choose a date and time that works best for your family, especially if you have young children.
- Avoid scheduling during meal times or nap times to ensure everyone is well-rested and in a good mood.
- Allow extra time for travel and preparation so you can arrive at the photoshoot location stress-free.
Pack Essentials
- Bring any props, outfits changes, or snacks that you may need during the photoshoot.
- Carry a small emergency kit with essentials like tissues, wet wipes, and a comb for quick touch-ups.
- Double-check the weather forecast and pack accordingly to stay comfortable during the shoot.
4. Relax and Have Fun
Be Yourself
- Encourage your family to relax and be themselves during the photoshoot to capture genuine expressions and emotions.
- Avoid forcing smiles or poses and instead focus on enjoying each other's company.
- Embrace the candid moments and let the photographer capture the natural interactions between family members.
Engage with Each Other
- Interact with your family members during the shoot by playing games, sharing jokes, or simply talking to each other.
- Encourage physical touch and closeness to create heartfelt and loving images that reflect your family bond.
- Trust the photographer to guide you through poses and activities that will result in beautiful and authentic photos.
5. Review and Preserve Your Photos
Select and Print Your Favorites
- Review the photos with your family and choose your favorite images for prints or digital copies.
- Consider creating a photo album, canvas prints, or framed photos to display in your home and cherish the memories.
- Share the photos with extended family members to spread the joy and celebrate special moments together.
Back Up Your Photos
- Store your digital photos in multiple locations, such as an external hard drive, cloud storage, or a USB drive, to prevent loss or damage.
- Organize your photos into folders labeled by date or event to easily locate and revisit your favorite family moments.
- Make regular backups of your photos to ensure that your precious memories are safe and secure for years to come.
By following these tips, you can make your professional family photoshoot a success and create picture-perfect memories that will be cherished for generations. Remember to relax, have fun, and enjoy the experience of capturing your family's unique bond through beautiful photography.
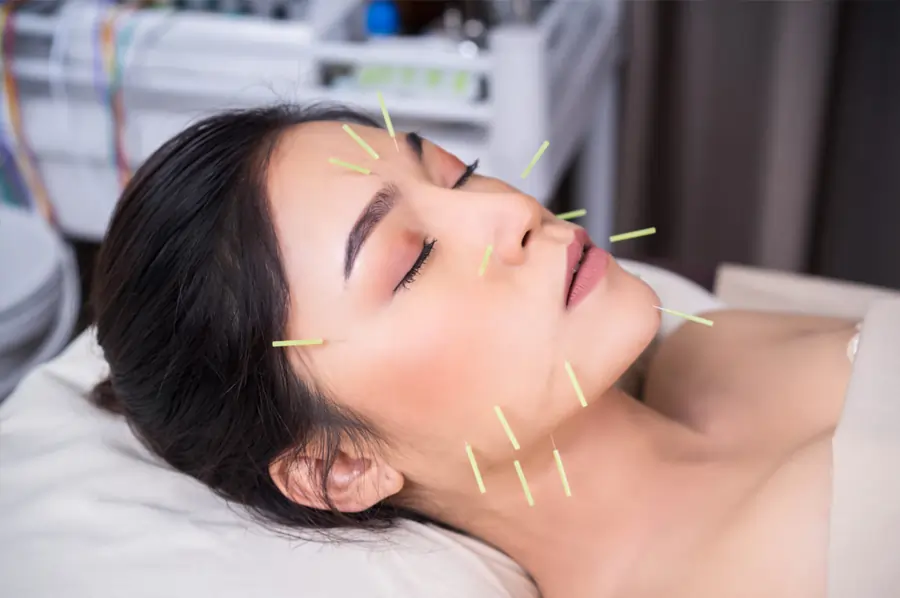
The Art of Facial Rejuvenation Acupuncture: Enhancing Your Beauty from Within
Facial rejuvenation acupuncture is a natural and non-invasive way to enhance your beauty by addressing both the physical and energetic aspects that contribute to aging. This ancient practice, also known as cosmetic acupuncture or acupuncture facelift, has been gaining popularity as a holistic alternative to invasive cosmetic procedures. By stimulating specific points on the face and body, facial rejuvenation acupuncture can help improve skin tone, reduce fine lines and wrinkles, and promote a healthy glow from within.
The Benefits of Facial Rejuvenation Acupuncture
Facial rejuvenation acupuncture offers a wide range of benefits for both the skin and overall well-being. Some of the key advantages of this holistic approach to beauty enhancement include:
- Reduction of fine lines and wrinkles
- Improved skin tone and texture
- Increased collagen production
- Enhanced muscle tone and firmness
- Stress reduction and relaxation
- Improved circulation for a healthy complexion
How Facial Rejuvenation Acupuncture Works
Facial rejuvenation acupuncture works by stimulating specific points on the face, neck, and body to improve the flow of Qi (life force energy) and blood circulation. This increased circulation helps nourish the skin and promote collagen production, which is essential for maintaining a youthful and radiant appearance. By addressing imbalances in the body's energy meridians, acupuncture can help restore harmony and vitality to the skin, leading to a more youthful and rejuvenated complexion.
The Process of Facial Rejuvenation Acupuncture
The process of facial rejuvenation acupuncture typically involves a series of weekly treatments that last around 60-90 minutes each. During a session, fine acupuncture needles are gently inserted into specific points on the face, neck, and body to stimulate the flow of energy and promote healing. The number of treatments required may vary depending on individual needs, but most people experience visible improvements in their skin after just a few sessions.
- Consultation: The acupuncturist will assess your skin condition and overall health to create a personalized treatment plan.
- Cleansing: The face is cleansed and prepped before the acupuncture treatment begins.
- Acupuncture: Fine needles are inserted into specific points on the face, neck, and body to stimulate energy flow and promote skin rejuvenation.
- Additional Therapies: Some practitioners may incorporate additional therapies such as facial massage, gua sha, or microcurrent stimulation to enhance the results.
- Follow-Up Care: It is essential to follow any post-treatment care instructions provided by your acupuncturist to maximize the benefits of the treatment.
Who Can Benefit from Facial Rejuvenation Acupuncture?
Facial rejuvenation acupuncture is suitable for individuals of all ages and skin types who are looking for a natural and holistic approach to beauty enhancement. This treatment can be particularly beneficial for those experiencing the following concerns:
- Fine lines and wrinkles
- Sagging skin
- Uneven skin tone
- Acne or rosacea
- Dull or tired-looking skin
- Stress-related skin issues
The Importance of Internal Health
Facial rejuvenation acupuncture goes beyond just treating the external signs of aging by addressing the underlying imbalances that contribute to skin issues. By promoting harmony within the body's energy system, acupuncture can help improve overall health and well-being, which is reflected in the skin's appearance. Emphasizing internal health is a key aspect of facial rejuvenation acupuncture, as true beauty comes from a healthy body, mind, and spirit.
Combining Acupuncture with Skincare
To enhance the results of facial rejuvenation acupuncture, it is important to complement the treatment with a good skincare routine and healthy lifestyle habits. Some tips for maintaining healthy and youthful skin in conjunction with acupuncture include:
- Stay hydrated by drinking plenty of water
- Eat a balanced diet rich in fruits, vegetables, and antioxidants
- Protect your skin from the sun with SPF and sun-protective clothing
- Avoid smoking and excessive alcohol consumption
- Practice stress-reducing activities such as meditation, yoga, or deep breathing exercises
Final Thoughts
Facial rejuvenation acupuncture offers a natural and holistic approach to enhancing your beauty from within. By addressing both the physical and energetic aspects of aging, this ancient practice can help you achieve a radiant and youthful complexion. Whether you are looking to reduce fine lines and wrinkles, improve skin tone, or simply relax and rejuvenate, facial rejuvenation acupuncture may be the perfect solution for you. Embrace the art of facial rejuvenation acupuncture and discover the transformative power of this age-old practice.
Pamper Yourself and Your Little One: Unboxing the Best Mommy-to-Be Box Options

As an expecting mom, self-care is essential not only for your well-being but also for the health of your growing baby. Mommy-to-be subscription boxes are a great way to treat yourself to some much-needed pampering and prepare for the arrival of your little one. Refer link: https://rumbly.co/
These curated boxes are filled with products specifically tailored to the needs of pregnant women, helping you feel relaxed, beautiful, and prepared during this special time. Let's explore some of the best mommy-to-be box options available.
1. What to Look for in a Mommy-to-Be Box
When choosing a mommy-to-be subscription box, it's important to consider what you need and what will make you feel most pampered. Here are some factors to keep in mind:
Key Considerations:
- Quality of products: Look for boxes that contain high-quality, pregnancy-safe products.
- Variety: Choose a box that offers a good mix of skincare, wellness, and baby-related products.
- Customization: Some boxes allow you to personalize your selections based on your preferences.
- Value for money: Consider the overall value of the box compared to its cost.
- Reviews: Read reviews from other expecting moms to get an idea of the box's quality and value.
2. Best Mommy-to-Be Box Options
a. Bump Boxes
Bump Boxes is a popular subscription service that offers monthly boxes filled with pregnancy-safe products tailored to each stage of your pregnancy. Each box includes a mix of skincare items, wellness products, and lifestyle goodies to help you feel your best during this special time.
b. Ecocentric Mom
Ecocentric Mom is a great option for eco-conscious moms-to-be. Their boxes feature organic, natural, and eco-friendly products for both mom and baby. From skincare items to baby accessories, Ecocentric Mom ensures that you and your little one are exposed to only the best, safest products.
c. Mama Bird Box
Mama Bird Box is a monthly subscription service that focuses on self-care for expecting moms. Their boxes include items such as bath salts, herbal teas, skincare products, and more. Mama Bird Box is all about helping you relax, unwind, and enjoy your pregnancy journey.
d. Oh Baby Boxes
Oh Baby Boxes offers a mix of pampering products for mom and practical items for baby. Their boxes include skincare products, snacks, baby gear, and other essentials to help you feel prepared for your little one's arrival. Oh Baby Boxes is a great way to treat yourself and start nesting for your baby.
3. Benefits of Mommy-to-Be Subscription Boxes
Signing up for a mommy-to-be subscription box can offer a variety of benefits for expecting moms:
Benefits Include:
- Convenience: Receive a curated selection of products delivered right to your door each month.
- Discover new products: Try out pregnancy-safe items that you may not have known about otherwise.
- Self-care reminder: Take time for yourself and prioritize self-care with a box designed just for you.
- Specialized items: Get products specifically chosen to support you during pregnancy and prepare for your baby.
- Excitement: Unboxing a surprise package each month can bring joy and anticipation to your pregnancy journey.
Whether you're looking to treat yourself, stock up on essentials, or discover new products, a mommy-to-be subscription box can be a fun and practical addition to your pregnancy experience. Take the time to explore different options and choose the box that best fits your needs and preferences.
The Future of Philanthropy: AI Grant Writers Making an Impact
The Future of Philanthropy: AI Grant Writers Making an Impact
In the dynamic field of philanthropy, securing grants is essential for organizations aiming to positively impact society. The traditional grant proposal writing process is often time-consuming and labor-intensive. Enter AI grant writers, a revolutionary force in fundraising. These technological solutions streamline the grant writing process, freeing organizations to focus more on their core missions rather than administrative tasks.
The Rise of AI Grant Writers
AI grant writers employ natural language processing and machine learning to parse grant guidelines, research funders, and craft proposals. By automating the search and application process, these AI systems enable organizations to quickly pinpoint and apply for relevant funding opportunities, significantly saving time and resources. For more insights on this technology, consider visiting ai grant writer.
Benefits of AI Grant Writers
- Efficiency: AI grant writers produce grant proposals much faster than humans, allowing nonprofits to allocate more time to other vital activities.
- Accuracy: These systems ensure proposals adhere to all required criteria, minimizing errors.
- Access to Opportunities: AI grant writers scan numerous funding sources to find grants that match an organizations mission and goals.
- Cost-Effectiveness: Employing AI grant writers can be more economical than hiring additional grant writing staff.
The Impact of AI Grant Writers on Philanthropy
The advent of AI grant writers is transforming the philanthropic landscape, optimizing grant-seeking processes and enhancing the potential for securing crucial funding. This shift not only drives innovation but also fosters significant change across various sectors, such as healthcare, education, and environmental conservation.
Key Contributions of AI Grant Writers
- Supporting Innovation: AI grant writers facilitate access to the funds necessary for pioneering solutions to social challenges.
- Enhancing Impact: By streamlining the grant application process, AI helps organizations better execute their programs, thereby amplifying their societal impact.
- Promoting Transparency: These tools ensure proposals align with funder priorities, enhancing transparency in the grant-making process.
- Fostering Collaboration: AI technology aids in identifying common funding opportunities and potential partnerships among organizations.
Challenges and Considerations
While AI grant writers offer substantial benefits, several challenges must be considered, such as potential biases in algorithm recommendations and the need for human oversight to ensure proposals reflect the organizations values. Furthermore, issues related to data security and the need for staff training are crucial when integrating AI into existing systems.
Considerations
- Alignment with Mission: It’s vital that AI grant writers resonate with an organization’s core values to maintain authenticity.
- Scalability: Organizations should evaluate the scalability of AI solutions to support growth and adapt to changing needs.
- Ethical Use of AI: Emphasizing ethical practices in AI usage is crucial to ensure fairness and accountability.
- Evaluation and Feedback: Regular assessment and stakeholder feedback are essential to refine and improve AI grant writing processes.
In conclusion, AI grant writers are poised to redefine philanthropy by enhancing the efficiency and effectiveness of grant-seeking endeavors. Despite potential challenges, the advantages of adopting AI grant writing technology are compelling, offering a significant boost to organizations striving to make a meaningful difference in the world.
The Art of Engaging Social Media Content: Tips and Tricks
Image Source- Google
Social media has become an integral part of our daily lives, and creating engaging content is key to standing out in a crowded digital landscape. Whether you're a business trying to reach new customers or an influencer looking to grow your following, mastering the art of creating content that captivates your audience is essential. You may browse through this website for the comprehensive range of offerings tailored to enhance your social media presence.
In this article, we will explore some tips and tricks to help you create social media content that not only grabs attention but also drives engagement and interaction.
Understanding Your Audience
One of the first steps in creating engaging social media content is to understand your audience. By knowing who you are trying to reach, you can tailor your content to their interests, preferences, and behaviors. Here are some tips for understanding your audience:
Research Your Audience
- Use analytics tools to gather data on your audience demographics, interests, and online behavior.
- Engage with your audience through surveys, polls, and comments to gather feedback and insights.
- Monitor trends and discussions within your industry to stay informed about what topics resonate with your audience.
Create Buyer Personas
- Develop detailed buyer personas that represent your target audience segments, including their demographics, interests, pain points, and goals.
- Use these buyer personas to guide your content creation process and ensure that your content is relevant and valuable to your audience.
Creating Compelling Visuals
Visual content is key to capturing your audience's attention on social media platforms. By creating compelling visuals that stand out in a sea of content, you can increase engagement and drive traffic to your profiles. Here are some tips for creating compelling visuals:
Use High-Quality Images and Videos
- Invest in high-quality images and videos that are clear, professional, and visually appealing.
- Optimize your visuals for each social media platform's specifications to ensure they display correctly and attract attention.
Add Graphic Elements
- Enhance your visuals with graphic elements such as text overlays, icons, and illustrations to make them more engaging and informative.
- Experiment with different graphic styles to see what resonates with your audience and drives the most engagement.
Writing Captivating Copy
In addition to compelling visuals, well-crafted copy is essential for capturing your audience's attention and conveying your message effectively. Here are some tips for writing captivating copy:
Craft Clear and Concise Headlines
- Create headlines that are concise, compelling, and relevant to your content to grab your audience's attention.
- Use power words, numbers, and questions to make your headlines more enticing and encourage clicks.
Include a Call to Action
- Include a clear call to action in your copy to prompt your audience to engage with your content, such as liking, sharing, commenting, or visiting your website.
- Make your call to action specific, actionable, and easy to follow to encourage higher engagement rates.
Encouraging Engagement
Engagement is the lifeblood of social media, and encouraging your audience to interact with your content is crucial for building a loyal following and driving traffic to your profiles. Here are some tips for encouraging engagement:
Ask Questions
- Encourage your audience to participate by asking questions in your posts and captions to spark conversations and gather feedback.
- Respond to comments and messages promptly to show that you value your audience's input and are actively listening to their feedback.
Run Contests and Giveaways
- Organize contests and giveaways to incentivize your audience to engage with your content and share it with their networks.
- Set clear rules and deadlines for your contests to create a sense of urgency and excitement among your audience.
Measuring Success
Finally, it's essential to measure the success of your social media content to understand what is working well and what can be improved. By analyzing key metrics and performance indicators, you can refine your content strategy and continue to create engaging content that resonates with your audience. Here are some tips for measuring success:
Track Key Performance Indicators
- Monitor key metrics such as likes, shares, comments, clicks, and conversions to gauge the performance of your social media content.
- Use analytics tools to track the reach, engagement, and ROI of your content and identify trends and patterns over time.
Experiment and Iterate
- Experiment with different types of content, posting times, and messaging to see what resonates best with your audience and drives the most engagement.
- Use A/B testing to compare different variations of your content and optimize your strategy based on the results.
By following these tips and tricks, you can master the art of creating engaging social media content that captivates your audience, drives interaction, and helps you achieve your goals on social media platforms. Remember to stay authentic, consistent, and responsive to your audience's needs and preferences to build a strong online presence and connect with your followers effectively.
Breaking Free: The Journey to Recovery at a Cannabis Rehab Center
Recovery from cannabis addiction is a journey that requires dedication, support, and professional guidance. Cannabis rehab centers provide a safe and structured environment for individuals looking to break free from their addiction and reclaim their lives. In this article, we will explore the process of recovery at a cannabis rehab center, the support and resources available, and the steps involved in overcoming addiction.
The Decision to Seek Help
Making the decision to seek help for cannabis addiction is the first step towards recovery. It takes courage and self-awareness to acknowledge that you need support to overcome your addiction. When you choose to enter a cannabis rehab center, you are taking a proactive step towards a healthier and more fulfilling life.
Signs that it's time to seek help:
- Increased tolerance to cannabis, needing more to achieve the same effects
- Withdrawal symptoms when not using cannabis
- Neglecting responsibilities and hobbies in favor of using cannabis
- Relationship problems due to cannabis use
Benefits of seeking help:
- Professional guidance and support
- A safe and structured environment for recovery
- Access to therapy and counseling services
- Opportunities for personal growth and self-discovery
Entering a Cannabis Rehab Center
Once you have made the decision to seek help, the next step is to enter a cannabis rehab center. This can be a daunting process, but remember that you are taking an important step towards a better future. Rehab centers offer a range of programs and services tailored to the needs of individuals struggling with cannabis addiction.
What to expect when entering a cannabis rehab center:
- Initial assessment to determine your treatment needs
- Detoxification process to rid your body of cannabis toxins
- Individual and group therapy sessions
- Educational workshops on addiction and recovery
- Healthy activities and coping mechanisms to replace cannabis use
The importance of a supportive environment:
Rehab centers provide a supportive environment where you can connect with others who are on a similar journey to recovery. The sense of community and camaraderie can be invaluable in helping you stay motivated and focused on your goals.
Rebuilding a Life Without Cannabis
Recovery from cannabis addiction is not just about stopping the use of the drug; it is also about rebuilding your life and creating a future free from addiction. Rehab centers offer resources and support to help you navigate this process and make positive changes in all areas of your life.
Steps towards rebuilding your life:
- Developing healthy habits and routines
- Rebuilding relationships with loved ones
- Exploring new hobbies and interests
- Setting and achieving personal and professional goals
Support after rehab:
Recovery from cannabis addiction is an ongoing process, and it is important to continue seeking support even after completing a rehab program. Rehab centers often offer aftercare services, such as support groups and counseling, to help you stay on track and maintain your sobriety.
Conclusion
Breaking free from cannabis addiction is a challenging but rewarding journey. With the right support and resources, individuals can overcome their addiction, reclaim their lives, and build a brighter future. If you or a loved one is struggling with cannabis addiction, consider reaching out to a cannabis rehab center for help and guidance on the path to recovery.
Interactive Children’s Church Lessons: Engaging Activities to Teach Important Values

Children's church lessons are a crucial part of a child's spiritual growth and development. It is essential to engage children in interactive and fun activities that help them understand and internalize important values. Interactive lessons not only make learning more enjoyable but also help children retain the teachings better. Here are some engaging activities that can be incorporated into children's church lessons to teach important values effectively:
1. Storytelling Sessions
Storytelling is a powerful tool to convey important messages and values to children. It captures their imagination and helps them relate to the lessons on a personal level. Incorporating storytelling sessions into children's church lessons can make the learning experience more interactive and engaging.
Benefits of Storytelling Sessions:
- Helps children connect with the lesson emotionally
- Captures children's attention and makes learning fun
- Encourages creativity and imagination
- Aids in better retention of important values
2. Role-Playing Activities
Role-playing activities allow children to step into different roles and scenarios, enabling them to understand the perspectives of others and learn important values such as empathy, kindness, and cooperation. This interactive approach helps children develop crucial social and emotional skills while having fun.
Benefits of Role-Playing Activities:
- Promotes empathy and understanding of others' feelings
- Encourages teamwork and cooperation
- Enhances communication skills
- Provides a hands-on way to learn and internalize important values
3. Arts and Crafts Projects
Arts and crafts projects are a creative and hands-on way to teach important values in children's church lessons. Children can express themselves through art while learning about concepts like love, forgiveness, and gratitude. These projects not only engage children but also help them reflect on the values they are learning.
Benefits of Arts and Crafts Projects:
- Encourages creativity and self-expression
- Helps children visualize abstract concepts through art
- Promotes fine motor skills and coordination
- Provides a tangible reminder of the lesson learned
4. Interactive Games
Interactive games are a fun and engaging way to reinforce important values and lessons in children's church. Games can be tailored to teach specific values such as honesty, respect, and faith. By incorporating games into children's church lessons, educators can make learning enjoyable and memorable for children.
Benefits of Interactive Games:
- Makes learning fun and engaging
- Reinforces teamwork and sportsmanship
- Encourages critical thinking and problem-solving
- Provides a lively and interactive learning environment
5. Music and Dance Sessions
Music and dance sessions are a vibrant and energetic way to teach important values in children's church lessons. Children can express themselves through music and movement while internalizing concepts like love, joy, and faith. These sessions not only engage children physically but also emotionally and spiritually.
Benefits of Music and Dance Sessions:
- Enhances emotional expression and connection
- Encourages physical activity and coordination
- Creates a joyful and uplifting atmosphere
- Facilitates a holistic learning experience
Interactive children's church lessons play a vital role in instilling important values and teachings in children. By incorporating engaging activities such as storytelling, role-playing, arts and crafts, interactive games, and music and dance sessions, educators can create a dynamic and enriching learning environment for children. These activities not only make learning fun and interactive but also help children develop essential social, emotional, and spiritual skills that will benefit them throughout their lives.
Mental Health Matters: Top-Rated Therapists in Los Angeles
Mental health is a crucial aspect of overall well-being, and seeking therapy can be a pivotal step towards maintaining good mental health. In a bustling city like Los Angeles, finding the right therapist can be overwhelming due to the plethora of options available. To help you navigate through the choices, we have compiled a list of the top-rated therapists in Los Angeles who can provide you with the support and guidance you need.
1. Dr. Emily White, PhD
Dr. Emily White is a licensed clinical psychologist with over 15 years of experience in the field. She specializes in cognitive-behavioral therapy and has helped numerous clients overcome anxiety, depression, and trauma.
Key Highlights:
- Individual and group therapy sessions available
- Accepts various insurance plans
- Warm and empathetic approach to therapy
2. Sarah Jones, LCSW
Sarah Jones is a licensed clinical social worker known for her compassionate and client-centered approach to therapy. She has a diverse background in working with individuals from different cultural backgrounds and specializes in LGBTQ+ affirming therapy.
Key Highlights:
- Expertise in trauma-informed care
- Offers sliding scale fees for those with financial constraints
- Virtual therapy sessions available
3. Dr. Michael Patel, PsyD
Dr. Michael Patel is a licensed psychologist with a focus on mindfulness-based therapy. He integrates mindfulness techniques with traditional psychotherapy to help clients achieve a sense of balance and well-being.
Key Highlights:
- Specializes in stress management and burnout prevention
- Experienced in working with high-achieving professionals
- Flexible scheduling options
4. Rachel Chang, LMFT
Rachel Chang is a licensed marriage and family therapist with a passion for helping couples and families navigate relationship challenges. She employs a strength-based approach to therapy, focusing on empowering her clients to make positive changes in their lives.
Key Highlights:
- Couples counseling and family therapy services offered
- Utilizes evidence-based practices
- Convenient location in the heart of Los Angeles
5. Dr. Javier Rodriguez, PhD
Dr. Javier Rodriguez is a bilingual psychologist who provides therapy in both English and Spanish. He has expertise in treating mood disorders and specializes in helping clients build resilience and coping skills.
Key Highlights:
- Culturally sensitive approach to therapy
- Skilled in treating anxiety and depression
- Teletherapy options for remote sessions
Choosing the right therapist is a personal decision that depends on your specific needs and preferences. It's essential to research and reach out to therapists to find the best fit for you. Remember, seeking help is a sign of strength, and taking care of your mental health is a vital investment in your overall well-being.
Building Dreams: Innovative Construction Services for Every Project
When it comes to construction projects, having the right team and services can make all the difference in turning dreams into reality. From residential homes to commercial buildings, innovative construction services are essential for bringing unique and ambitious projects to life. Here, we will explore the various innovative construction services available for every type of project.

Residential Construction Services
New Home Construction
- Custom Home Design: Innovative construction services often include custom home design to meet the specific needs and style preferences of homeowners.
- Sustainable Building Practices: Builders are increasingly incorporating sustainable building practices into residential construction projects to reduce environmental impact and energy costs.
- Smart Home Technology Integration: Integrating smart home technology into new construction projects is becoming more common, offering homeowners greater convenience and energy efficiency.
Home Renovations and Additions
- Interior Upgrades: From kitchen remodels to bathroom renovations, innovative construction services can transform existing spaces to meet modern design trends and functional needs.
- Room Additions: Builders can seamlessly integrate new room additions into existing homes, expanding living space and increasing property value.
- Energy-Efficient Upgrades: Renovations often include energy-efficient upgrades such as windows, insulation, and HVAC systems to improve comfort and reduce utility costs.
Commercial Construction Services
Office Buildings
- Green Building Certification: Many innovative construction services offer expertise in obtaining green building certifications such as LEED to enhance sustainability and appeal to environmentally conscious tenants.
- Open Concept Layouts: Modern office buildings often feature open concept layouts that promote collaboration and flexibility, requiring innovative construction techniques to achieve.
- Advanced Technology Infrastructure: Commercial construction services can incorporate advanced technology infrastructure to support the connectivity and communication needs of modern businesses.
Retail Spaces
- Visual Merchandising Solutions: Builders can provide innovative solutions for visual merchandising to create attractive and engaging retail spaces that drive customer traffic and sales.
- Pop-Up Store Construction: Temporary pop-up stores require quick and efficient construction services to capitalize on short-term retail opportunities.
- Interactive Customer Experiences: Construction services can help retailers design and build interactive customer experiences such as digital displays and interactive kiosks.
Specialized Construction Services
Industrial Projects
- Heavy Equipment Foundations: Industrial construction services often involve the installation of specialized foundations to support heavy equipment and machinery.
- Warehousing Solutions: Builders can create innovative warehousing solutions that maximize storage capacity and operational efficiency for industrial clients.
- Industrial Safety Compliance: Construction services for industrial projects prioritize safety compliance to protect workers and ensure regulatory adherence.
Hospitality Construction
- Luxury Resort Development: Construction services for luxury resorts require expertise in high-end finishes, unique architectural features, and guest experience design.
- Restaurant Build-Outs: Builders can transform existing spaces into functional and stylish restaurants that cater to the needs of chefs, staff, and diners.
- Hotel Renovations: Renovations of existing hotels often include updates to guest rooms, common areas, and amenities to maintain competitiveness in the hospitality industry.
Conclusion
From custom home design to industrial projects, innovative construction services play a crucial role in bringing dreams to life across a variety of sectors. With sustainable practices, advanced technology integration, and a focus on quality and efficiency, construction services continue to evolve to meet the changing needs of clients and projects. By partnering with experienced builders and contractors who offer innovative solutions, every construction project has the potential to exceed expectations and deliver exceptional results.
Expert Home Cleaning Services for Busy Lifestyles

Keeping your home clean and organized is essential for a happy and healthy living environment. However, with busy work schedules and family commitments, finding the time to maintain a pristine home can be challenging. This is where professional home cleaning services come in to save the day. Expert cleaners can help you keep your home sparkling clean, allowing you to focus on other important aspects of your life.
The Benefits of Hiring Expert Home Cleaning Services
Convenience
- Save time and effort by outsourcing the cleaning tasks to professionals.
- Flexible scheduling options to fit your busy lifestyle.
- Have a clean home without lifting a finger.
Expertise
- Trained and experienced cleaners who know the best cleaning techniques and products.
- Attention to detail to ensure every corner of your home is spotless.
- Specialized cleaning services for different areas of your home, such as kitchens, bathrooms, and bedrooms.
Healthy Living Environment
- Regular cleaning helps eliminate dust, allergens, and bacteria from your home.
- Reduce the risk of illnesses and allergies by maintaining a clean living space.
- Breath easy knowing that your home is clean and sanitized.
Types of Cleaning Services Offered
Regular Cleaning
- Weekly, bi-weekly, or monthly cleaning services to keep your home consistently clean.
- Dusting, vacuuming, mopping, and bathroom cleaning included in regular cleaning packages.
- Customizable cleaning plans to suit your specific needs and preferences.
Deep Cleaning
- A thorough cleaning of your entire home, including hard-to-reach areas and neglected spaces.
- Perfect for spring cleaning or preparing for a special event or guests.
- Includes cleaning of inside cabinets, appliances, baseboards, and more.
Move-In/Move-Out Cleaning
- Ensure your new home is clean and sanitized before moving in.
- Get your security deposit back by leaving your previous home spotless.
- Cleaning of all surfaces, floors, and fixtures to make the transition smoother.
How to Choose the Right Home Cleaning Service
Ask for Recommendations
- Get referrals from friends, family, or neighbors who have used cleaning services before.
- Check online reviews and ratings to gauge the reputation of the cleaning company.
Inquire About Services and Pricing
- Ask about the types of cleaning services offered and whether they align with your needs.
- Get a detailed quote that includes all costs and any additional fees for special requests.
Consider the Company's Reputation and Experience
- Choose a cleaning service with a proven track record of customer satisfaction.
- Look for a company that has been in the business for several years and has experienced cleaners.
Check for Insurance and Bonding
- Ensure the cleaning company is properly insured and bonded to protect your home and belongings.
- Inquire about liability coverage in case of accidents or damages during the cleaning process.
Conclusion
Expert home cleaning services are a valuable resource for individuals with busy lifestyles who want to maintain a clean and healthy living environment. By outsourcing your cleaning tasks to professionals, you can enjoy a spotless home without the stress and hassle of doing it yourself. With a wide range of cleaning services available, you can choose the option that best fits your needs and schedule. Take the first step towards a cleaner home by hiring expert home cleaning services today!